Reveal the signals in your data with the biomedical analytics platform that cares about optimizing usability, out-of-the-box functionality, extensibility, and speed—all with the lowest cloud storage and computing costs.
Catalyze your research.
Securely. Collaboratively. Reproducibly.
Extensible application packages that scientists actually want to use, informed by years of close partnerships.
Visually probe complex multimodal longitudinal data sets. Or use science-level APIs that do the heavy lifting by hiding implementation details.
Seamlessly synchronize interactive work by GUI and notebook users.
Get cost relief for:
Sr. V.P., Data Science

Services
Accelerate your programs with P4’s Solution Services
We deliver as committed
Top 10 Pharma Program Manager for a new program: “Paradigm4 knocked this project out of the park and exceeded expectations on timeline and quality of work….you guys have been rock stars on a very challenging project.”
We’re productive
In the words of a Technical Product Manager: “I attend so many meetings every week where stuff is run into the ground. It is refreshing to attend these meetings [with P4] where work actually gets done.”
We’re responsive
Another top 20 pharma customer said:
“You are always quick to respond to issues we have and investigate and prioritize as they come up. You respond faster than other people inside our company.”
Our Team
Our Solutions team has extensive experience and advanced degrees in bioinformatics, biomedical engineering, population genetics, image and signal processing, applied mathematics, machine-learning, scalable parallel computing, R, Python, databases, and cloud computing.
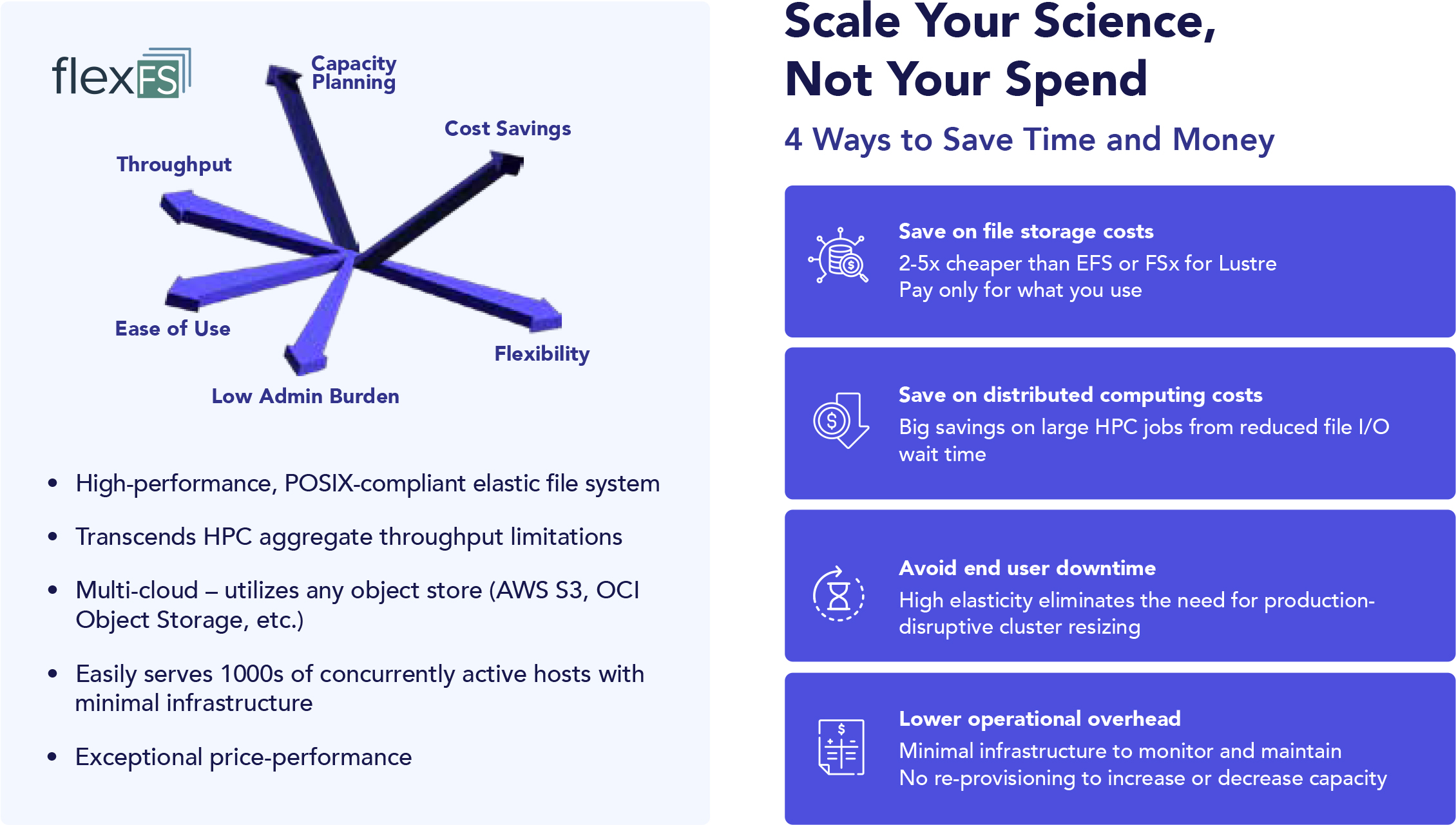
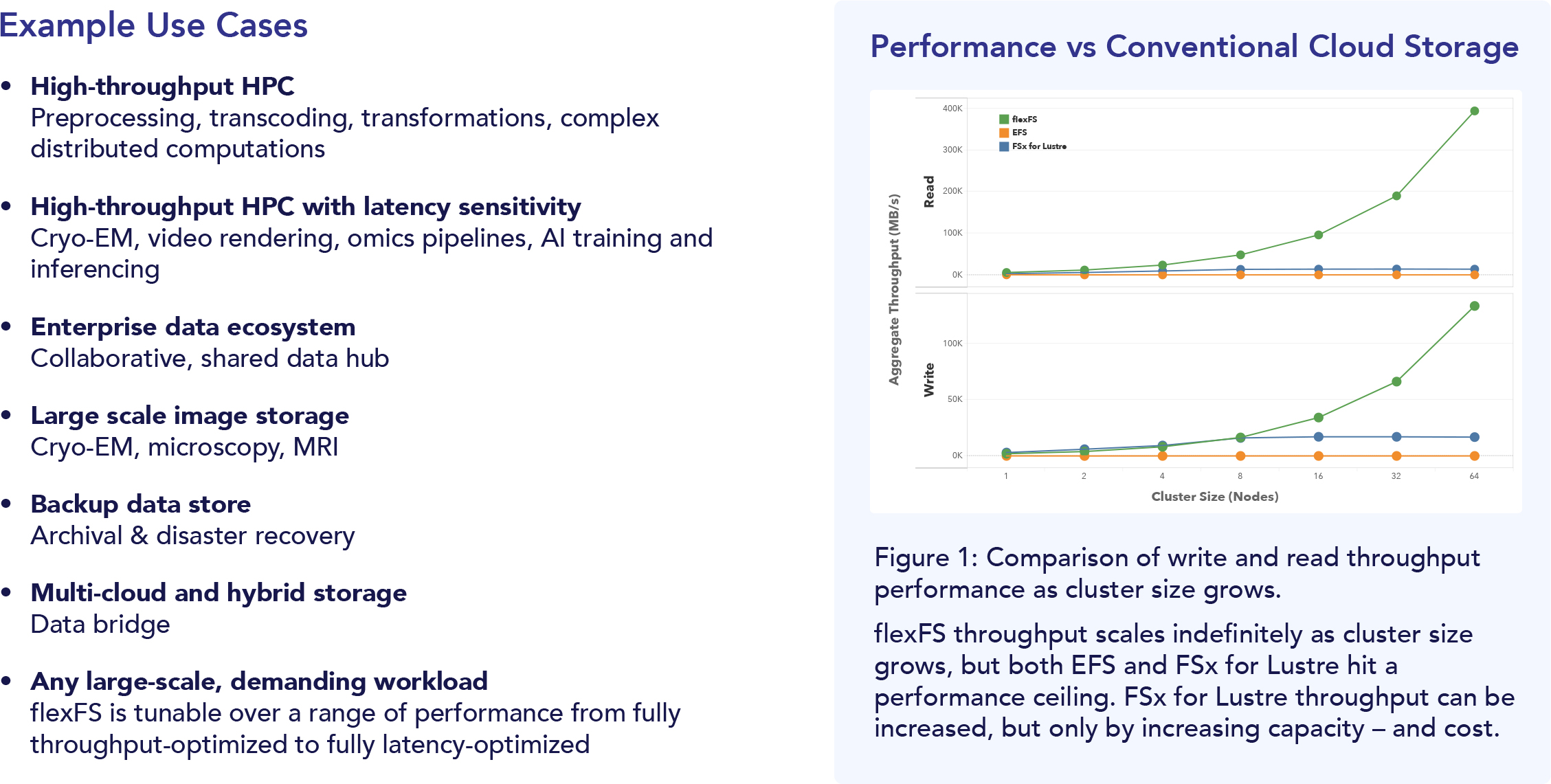

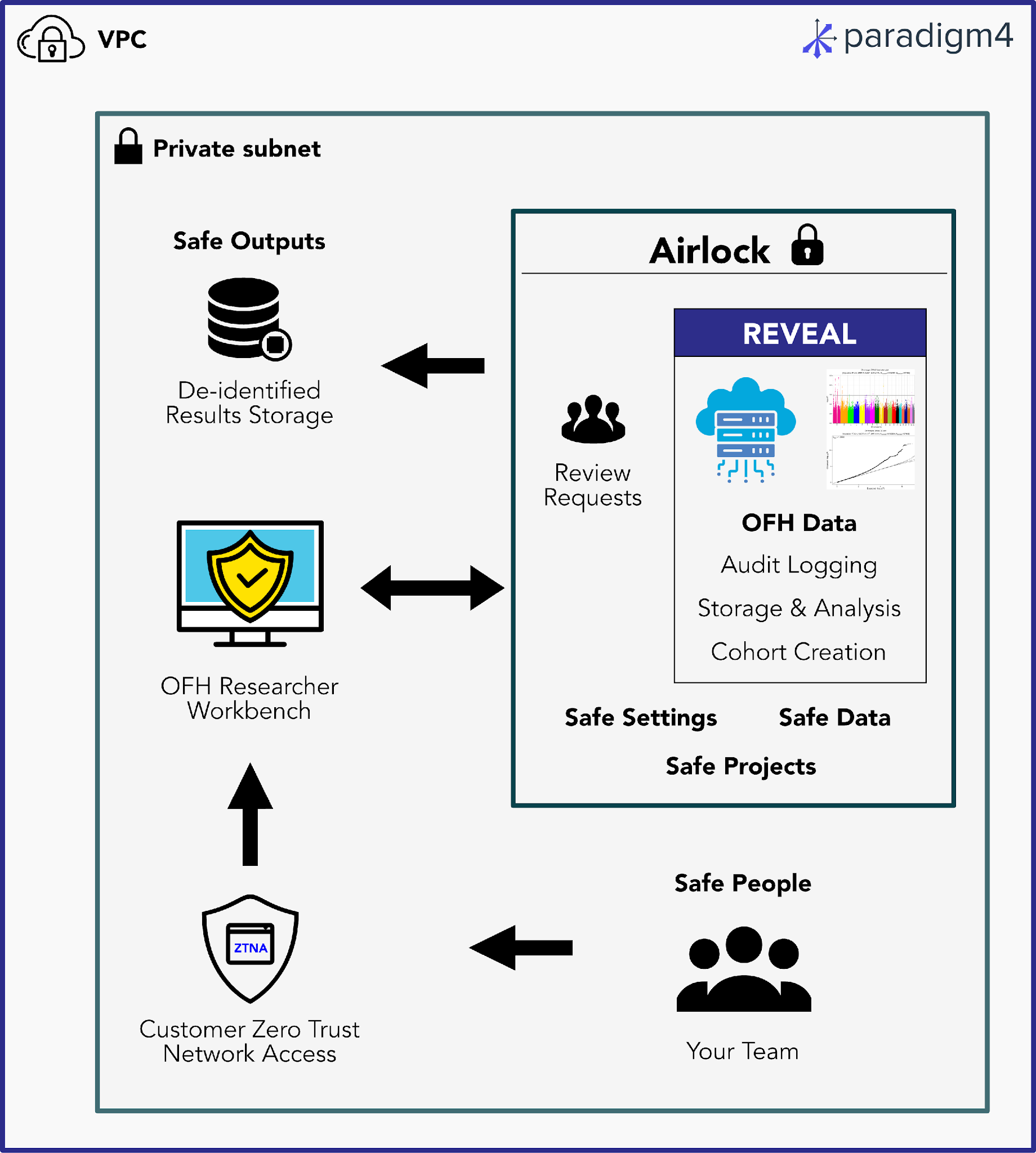
Solid security and governance are table stakes